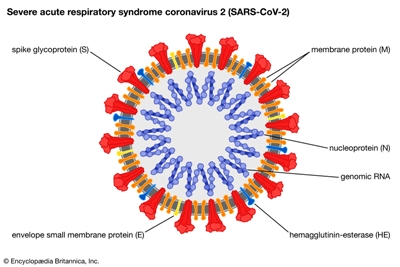
This model shows the structure of the spike protein in its closed configuration, in its original D614 form (left) and its mutant form (G614). In the mutant spike protein, the 630 loop (in red) stabilizes the spike, preventing it from flipping open prematurely and rendering SARS-CoV-2 more infectious. [Bing Chen, PhD, Boston Children's Hospital]
The SARS-CoV-2 variants that have emerged in the past few months have attracted an immense amount of speculation. Specifically, whether the mutations may confer an advantage to the virus, render our antibodies ineffective, and lead to vaccine escape. New research has uncovered why the G614 strain (with a substitution for aspartic acid by glycine at position 614 in the spike protein) and its recent variants—which are now the dominant circulating forms of the virus—facilitate rapid viral spread.
The work, led by Bing Chen, PhD, at Boston Children’s Hospital, analyzed how the structure of the spike protein changes with the D614G mutation. Using cryo-electron microscopy (cryo-EM), which has resolution down to the atomic level, they report structures of a full-length G614 spike trimer which adopts three distinct prefusion conformations differing primarily by the position of one receptor-binding domain (RBD). Chen’s team found that the D614G mutation makes the spike more stable as compared with the original SARS-CoV-2 virus.
This work is published in Science, in the paper, “Structural impact on SARS-CoV-2 spike protein by D614G substitution.”
In the original coronavirus, the spike proteins would bind to the ACE2 receptor and then dramatically change shape, folding in on themselves. This enabled the virus to fuse its membrane with our own cells’ membranes and get inside. However, as Chen and colleagues reported in July 2020, the spikes would sometimes . . .